Titanic Case Analysis
Titanic: Machine Learning from Disaster
Predict survival on the Titanic
- Defining the problem statement
- Collecting the data
- Exploratory data analysis
- Feature engineering
- Modelling
- Testing
1. Defining the problem statement:
Conducting an analysis on predicting the sort of people who were likely to survive. We are going to apply the tools of machine learning to predict which passengers survivied the tragedy
from IPython.display import Image
Image(url= "https://static1.squarespace.com/static/5006453fe4b09ef2252ba068/5095eabce4b06cb305058603/5095eabce4b02d37bef4c24c/1352002236895/100_anniversary_titanic_sinking_by_esai8mellows-d4xbme8.jpg")

2. Collecting the Data:
The data comes from Kaggle, they are providing us with training and testing set. link to the data is found on my github: https://github.com/Haalibrahim/Titanic-case-study
Load libraries, train and test data
#Import Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set()
plt.style.use('fivethirtyeight')
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
train=pd.read_csv('train.csv')
test=pd.read_csv('test.csv')
3. Exploratory data analysis:
Data Dictionary:
- Survived: 0 = No, 1= Yes
- Pclass: Ticket class 1 = 1st, 2= 2nd, 3 = 3rd
- Sibsp: Number of siblings / spouses aboard the titanic
- Parch: Number of parents/ childern abroad the titanic
- Ticket: Ticket number
- Fare: price of the ticket
- Cabin: Number of the Cabin
- Embarked: Port of Embarkation, C= Cherbourg , Q= Queenstown, S= Southhampton
train.head(10)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
| 5 | 6 | 0 | 3 | Moran, Mr. James | male | NaN | 0 | 0 | 330877 | 8.4583 | NaN | Q |
| 6 | 7 | 0 | 1 | McCarthy, Mr. Timothy J | male | 54.0 | 0 | 0 | 17463 | 51.8625 | E46 | S |
| 7 | 8 | 0 | 3 | Palsson, Master. Gosta Leonard | male | 2.0 | 3 | 1 | 349909 | 21.0750 | NaN | S |
| 8 | 9 | 1 | 3 | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) | female | 27.0 | 0 | 2 | 347742 | 11.1333 | NaN | S |
| 9 | 10 | 1 | 2 | Nasser, Mrs. Nicholas (Adele Achem) | female | 14.0 | 1 | 0 | 237736 | 30.0708 | NaN | C |
test.head()
| PassengerId | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 892 | 3 | Kelly, Mr. James | male | 34.5 | 0 | 0 | 330911 | 7.8292 | NaN | Q |
| 1 | 893 | 3 | Wilkes, Mrs. James (Ellen Needs) | female | 47.0 | 1 | 0 | 363272 | 7.0000 | NaN | S |
| 2 | 894 | 2 | Myles, Mr. Thomas Francis | male | 62.0 | 0 | 0 | 240276 | 9.6875 | NaN | Q |
| 3 | 895 | 3 | Wirz, Mr. Albert | male | 27.0 | 0 | 0 | 315154 | 8.6625 | NaN | S |
| 4 | 896 | 3 | Hirvonen, Mrs. Alexander (Helga E Lindqvist) | female | 22.0 | 1 | 1 | 3101298 | 12.2875 | NaN | S |
train.shape
(891, 12)
test.shape
(418, 11)
train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
PassengerId 891 non-null int64
Survived 891 non-null int64
Pclass 891 non-null int64
Name 891 non-null object
Sex 891 non-null object
Age 714 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Ticket 891 non-null object
Fare 891 non-null float64
Cabin 204 non-null object
Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
test.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 418 entries, 0 to 417
Data columns (total 11 columns):
PassengerId 418 non-null int64
Pclass 418 non-null int64
Name 418 non-null object
Sex 418 non-null object
Age 332 non-null float64
SibSp 418 non-null int64
Parch 418 non-null int64
Ticket 418 non-null object
Fare 417 non-null float64
Cabin 91 non-null object
Embarked 418 non-null object
dtypes: float64(2), int64(4), object(5)
memory usage: 36.0+ KB
train.isna().sum() # = train.isnull().sum()
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
test.isna().sum()
PassengerId 0
Pclass 0
Name 0
Sex 0
Age 86
SibSp 0
Parch 0
Ticket 0
Fare 1
Cabin 327
Embarked 0
dtype: int64
#Let us explore the data: (pay attention to the age column)
train.describe()
| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| count | 891.000000 | 891.000000 | 891.000000 | 714.000000 | 891.000000 | 891.000000 | 891.000000 |
| mean | 446.000000 | 0.383838 | 2.308642 | 29.699118 | 0.523008 | 0.381594 | 32.204208 |
| std | 257.353842 | 0.486592 | 0.836071 | 14.526497 | 1.102743 | 0.806057 | 49.693429 |
| min | 1.000000 | 0.000000 | 1.000000 | 0.420000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 223.500000 | 0.000000 | 2.000000 | 20.125000 | 0.000000 | 0.000000 | 7.910400 |
| 50% | 446.000000 | 0.000000 | 3.000000 | 28.000000 | 0.000000 | 0.000000 | 14.454200 |
| 75% | 668.500000 | 1.000000 | 3.000000 | 38.000000 | 1.000000 | 0.000000 | 31.000000 |
| max | 891.000000 | 1.000000 | 3.000000 | 80.000000 | 8.000000 | 6.000000 | 512.329200 |
test.describe()
| PassengerId | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|
| count | 418.000000 | 418.000000 | 332.000000 | 418.000000 | 418.000000 | 417.000000 |
| mean | 1100.500000 | 2.265550 | 30.272590 | 0.447368 | 0.392344 | 35.627188 |
| std | 120.810458 | 0.841838 | 14.181209 | 0.896760 | 0.981429 | 55.907576 |
| min | 892.000000 | 1.000000 | 0.170000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 996.250000 | 1.000000 | 21.000000 | 0.000000 | 0.000000 | 7.895800 |
| 50% | 1100.500000 | 3.000000 | 27.000000 | 0.000000 | 0.000000 | 14.454200 |
| 75% | 1204.750000 | 3.000000 | 39.000000 | 1.000000 | 0.000000 | 31.500000 |
| max | 1309.000000 | 3.000000 | 76.000000 | 8.000000 | 9.000000 | 512.329200 |
Bar Chart for Categorical Features
- Pclass
- Sex
- SibSp ( # of siblings and spouse)
- Parch ( # of parents and children)
- Embarked
- Cabin
# Count of people who survive:
train['Survived'].value_counts()
0 549
1 342
Name: Survived, dtype: int64
def bar_chart(feature):
Survived= train[train['Survived']==1][feature].value_counts()
Dead= train[train['Survived']==0][feature].value_counts()
df = pd.DataFrame([Survived,Dead])
df.index= ['Survived','Dead']
df.plot(kind='bar', stacked =True, figsize =(10,5))
bar_chart('Sex')

Women are more likely to survive than Men
bar_chart('Pclass')

The Chart confirms 1st class more likely survivied than other classes The Chart confirms 3rd class more likely dead than other classes
bar_chart('SibSp')

The Chart confirms a person aboarded with more than 2 siblings or spouse more likely survived The Chart confirms a person aboarded without siblings or spouse more likely dead
bar_chart('Parch')

The Chart confirms a person aboarded with more than 2 parents or children more likely survived The Chart confirms a person aboarded alone more likely dead
bar_chart('Embarked')

- The Chart confirms a person aboarded from C slightly more likely survived
- The Chart confirms a person aboarded from Q more likely dead
- The Chart confirms a person aboarded from S more likely dead
4. Feature engineering
- The purpose of Feautre engineering is to prepare the data for machine learning algorithms by creating feature vectors.
- Feature vector is an n dimensional vector that represent some object
train.head()
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
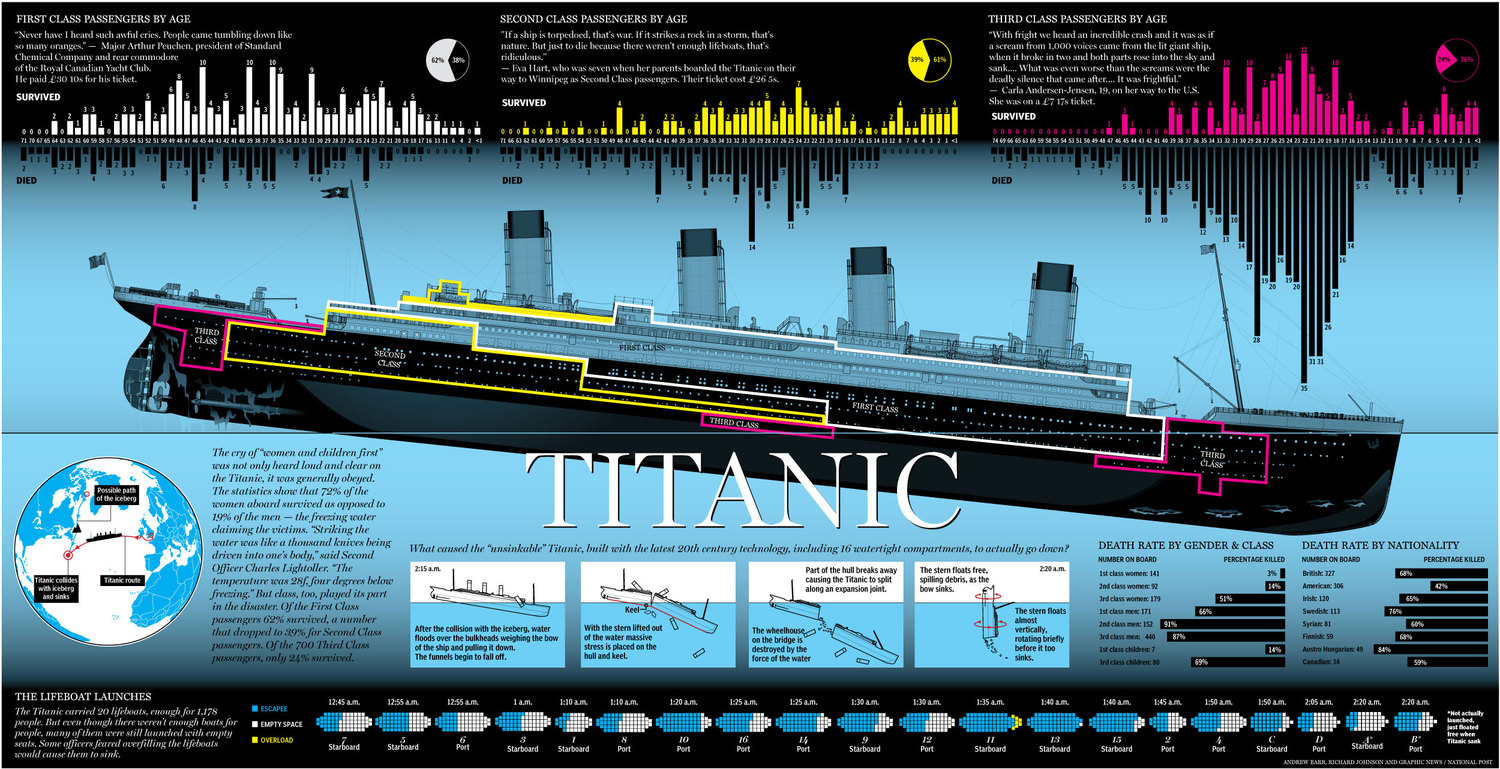
4.1 how titanic sank?
- sank from the bow of the ship where third class rooms located
- conclusion, Pclass is key feature for classifier
Image(url= "https://static1.squarespace.com/static/5006453fe4b09ef2252ba068/t/5090b249e4b047ba54dfd258/1351660113175/TItanic-Survival-Infographic.jpg?format=1500w")
4.2 Name
train_test_data=[train,test] # combining train and test dataset
train['Title']=0
for i in train_test_data:
train['Title']=train.Name.str.extract('([A-Za-z]+)\.') #extracting Name initials
test['Title']=test.Name.str.extract('([A-Za-z]+)\.')
train['Title'].value_counts()
Mr 517
Miss 182
Mrs 125
Master 40
Dr 7
Rev 6
Mlle 2
Major 2
Col 2
Jonkheer 1
Don 1
Lady 1
Sir 1
Mme 1
Ms 1
Capt 1
Countess 1
Name: Title, dtype: int64
train['Title'].value_counts().sum()
891
test['Title'].value_counts()
Mr 240
Miss 78
Mrs 72
Master 21
Rev 2
Col 2
Dr 1
Dona 1
Ms 1
Name: Title, dtype: int64
test['Title'].value_counts().sum()
418
pd.crosstab(train.Title,train.Sex).T.style.background_gradient(cmap='summer_r')
| Title | Capt | Col | Countess | Don | Dr | Jonkheer | Lady | Major | Master | Miss | Mlle | Mme | Mr | Mrs | Ms | Rev | Sir |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||||||||||
| female | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 182 | 2 | 1 | 0 | 125 | 1 | 0 | 0 |
| male | 1 | 2 | 0 | 1 | 6 | 1 | 0 | 2 | 40 | 0 | 0 | 0 | 517 | 0 | 0 | 6 | 1 |
pd.crosstab(test.Title,test.Sex).T.style.background_gradient(cmap='summer_r')
| Title | Col | Dona | Dr | Master | Miss | Mr | Mrs | Ms | Rev |
|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||
| female | 0 | 1 | 0 | 0 | 78 | 0 | 72 | 1 | 0 |
| male | 2 | 0 | 1 | 21 | 0 | 240 | 0 | 0 | 2 |
train['Title'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess',
'Jonkheer','Col','Rev','Capt','Sir','Don'],['Miss',
'Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr'],inplace=True)
test['Title'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess',
'Jonkheer','Col','Rev','Capt','Sir','Don'],['Miss',
'Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr'],inplace=True)
train.head()
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | Mr |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | Mrs |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | Miss |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | Mrs |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | Mr |
train['Title'].value_counts()
Mr 529
Miss 186
Mrs 127
Master 40
Other 9
Name: Title, dtype: int64
train['Title'].value_counts().sum()
891
test['Title'].value_counts()
Mr 241
Miss 79
Mrs 72
Master 21
Other 4
Dona 1
Name: Title, dtype: int64
test['Title'].value_counts().sum()
418
# delete unnecessary feature from dataset
train.drop('Name', axis=1, inplace=True)
test.drop('Name', axis=1, inplace=True)
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | Mr |
| 1 | 2 | 1 | 1 | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | Mrs |
| 2 | 3 | 1 | 3 | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | Miss |
| 3 | 4 | 1 | 1 | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | Mrs |
| 4 | 5 | 0 | 3 | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | Mr |
test.head()
| PassengerId | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 892 | 3 | male | 34.5 | 0 | 0 | 330911 | 7.8292 | NaN | Q | Mr |
| 1 | 893 | 3 | female | 47.0 | 1 | 0 | 363272 | 7.0000 | NaN | S | Mrs |
| 2 | 894 | 2 | male | 62.0 | 0 | 0 | 240276 | 9.6875 | NaN | Q | Mr |
| 3 | 895 | 3 | male | 27.0 | 0 | 0 | 315154 | 8.6625 | NaN | S | Mr |
| 4 | 896 | 3 | female | 22.0 | 1 | 1 | 3101298 | 12.2875 | NaN | S | Mrs |
pd.crosstab(train.Title,train.Sex).T.style.background_gradient(cmap='summer_r')
| Title | Master | Miss | Mr | Mrs | Other |
|---|---|---|---|---|---|
| Sex | |||||
| female | 0 | 186 | 1 | 127 | 0 |
| male | 40 | 0 | 528 | 0 | 9 |
bar_chart('Title')

4.3 Age
print('Oldest person Survived was of:',train['Age'].max())
print('Youngest person Survived was of:',train['Age'].min())
print('Average person Survived was of:',train['Age'].mean())
Oldest person Survived was of: 80.0
Youngest person Survived was of: 0.42
Average person Survived was of: 29.69911764705882
f,ax=plt.subplots(1,2,figsize=(18,8))
sns.violinplot('Pclass','Age',hue='Survived',data=train,split=True,ax=ax[0])
ax[0].set_title('PClass and Age vs Survived')
ax[0].set_yticks(range(0,110,10))
sns.violinplot("Sex","Age", hue="Survived", data=train,split=True,ax=ax[1])
ax[1].set_title('Sex and Age vs Survived')
ax[1].set_yticks(range(0,110,10))
plt.show()

train.groupby('Title')['Age'].mean()
Title
Master 4.574167
Miss 21.860000
Mr 32.739609
Mrs 35.981818
Other 45.888889
Name: Age, dtype: float64
train.loc[(train.Age.isnull()) & (train.Title=='Mr'),'Age']=33
train.loc[(train.Age.isnull()) & (train.Title=='Mrs'),'Age']=36
train.loc[(train.Age.isnull()) & (train.Title=='Master'),'Age']=5
train.loc[(train.Age.isnull()) & (train.Title=='Miss'),'Age']=22
train.loc[(train.Age.isnull()) & (train.Title=='Other'),'Age']=46
train.Age.isnull().any()
False
Title map
- Mr : 0
- Miss : 1
- Mrs: 2
- Master: 3
- Other: 4
title_mapping = {"Mr": 0, "Miss": 1, "Mrs": 2,
"Master": 3, "Other":4 }
for dataset in train_test_data:
dataset['Title'] = dataset['Title'].map(title_mapping)
bar_chart('Title')

train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | 0 |
| 1 | 2 | 1 | 1 | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | 2 |
| 2 | 3 | 1 | 3 | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | 1 |
| 3 | 4 | 1 | 1 | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | 2 |
| 4 | 5 | 0 | 3 | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | 0 |
test.head()
| PassengerId | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 892 | 3 | male | 34.5 | 0 | 0 | 330911 | 7.8292 | NaN | Q | 0.0 |
| 1 | 893 | 3 | female | 47.0 | 1 | 0 | 363272 | 7.0000 | NaN | S | 2.0 |
| 2 | 894 | 2 | male | 62.0 | 0 | 0 | 240276 | 9.6875 | NaN | Q | 0.0 |
| 3 | 895 | 3 | male | 27.0 | 0 | 0 | 315154 | 8.6625 | NaN | S | 0.0 |
| 4 | 896 | 3 | female | 22.0 | 1 | 1 | 3101298 | 12.2875 | NaN | S | 2.0 |
f,ax=plt.subplots(1,2,figsize=(20,20))
train[train['Survived']==0].Age.plot.hist(ax=ax[0],bins=20,edgecolor='black',color='red')
ax[0].set_title('Survived = 0')
x1=list(range(0,85,5))
ax[0].set_xticks(x1)
train[train['Survived']==1].Age.plot.hist(ax=ax[1],bins=20,edgecolor='black',color='green')
x2=list(range(0,85,5))
ax[1].set_xticks(x2)
ax[1].set_title('Survived = 1')
plt.show()

facet =sns.FacetGrid(train, hue='Survived',aspect=4)
facet.map(sns.kdeplot,'Age',shade=True)
facet.set(xlim=(0, train['Age'].max()))
facet.add_legend()
plt.show()

grid = sns.FacetGrid(train, col='Survived', row='Pclass', size=2.2, aspect=1.6)
grid.map(plt.hist, 'Age', alpha=.5, bins=20)
grid.add_legend();

for dataset in train_test_data:
dataset.loc[dataset['Age']<= 16, 'Age']= 0,
dataset.loc[(dataset['Age']> 16) & (dataset['Age']<=26), 'Age']= 1,
dataset.loc[(dataset['Age']> 26) & (dataset['Age']<=36), 'Age']= 2,
dataset.loc[(dataset['Age']> 36) & (dataset['Age']<=62), 'Age']= 3,
dataset.loc[dataset['Age']> 62, 'Age']= 4
bar_chart("Age")

train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | male | 1.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | 0 |
| 1 | 2 | 1 | 1 | female | 3.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | 2 |
| 2 | 3 | 1 | 3 | female | 1.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | 1 |
| 3 | 4 | 1 | 1 | female | 2.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | 2 |
| 4 | 5 | 0 | 3 | male | 2.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | 0 |
4.4 Sex
train.groupby(['Sex', 'Survived'])['Survived'].count()
Sex Survived
female 0 81
1 233
male 0 468
1 109
Name: Survived, dtype: int64
train.groupby('Sex')[['Survived']].mean()
| Survived | |
|---|---|
| Sex | |
| female | 0.742038 |
| male | 0.188908 |
train[['Sex','Survived']].groupby(['Sex']).mean().plot.bar()
sns.countplot('Sex',hue='Survived',data=train,)
plt.show()

sex_mapping = {"male": 0, "female": 1}
for dataset in train_test_data:
dataset['Sex'] = dataset['Sex'].map(sex_mapping)
bar_chart('Sex')

train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | 0 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | 2 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | 1 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | 2 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | 0 |
train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
PassengerId 891 non-null int64
Survived 891 non-null int64
Pclass 891 non-null int64
Sex 891 non-null int64
Age 891 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Ticket 891 non-null object
Fare 891 non-null float64
Cabin 204 non-null object
Embarked 889 non-null object
Title 891 non-null int64
dtypes: float64(2), int64(7), object(3)
memory usage: 83.7+ KB
test.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 418 entries, 0 to 417
Data columns (total 11 columns):
PassengerId 418 non-null int64
Pclass 418 non-null int64
Sex 418 non-null int64
Age 332 non-null float64
SibSp 418 non-null int64
Parch 418 non-null int64
Ticket 418 non-null object
Fare 417 non-null float64
Cabin 91 non-null object
Embarked 418 non-null object
Title 417 non-null float64
dtypes: float64(3), int64(5), object(3)
memory usage: 36.0+ KB
4.5 Pclass
sns.countplot('Pclass', hue='Survived', data=train)
plt.title('Pclass: Sruvived vs Dead')
plt.show()

pd.crosstab([train.Sex,train.Survived],train.Pclass,margins=True).style.background_gradient(cmap='summer_r')
| Pclass | 1 | 2 | 3 | All | |
|---|---|---|---|---|---|
| Sex | Survived | ||||
| 0 | 0 | 77 | 91 | 300 | 468 |
| 1 | 45 | 17 | 47 | 109 | |
| 1 | 0 | 3 | 6 | 72 | 81 |
| 1 | 91 | 70 | 72 | 233 | |
| All | 216 | 184 | 491 | 891 |
sns.factorplot('Pclass', 'Survived', hue='Sex', data=train)
plt.show()

sns.factorplot('Pclass','Survived',col='Title',data=train)
plt.show()

4.6 SibSip
pd.crosstab([train.SibSp],train.Survived).style.background_gradient('summer_r')
| Survived | 0 | 1 |
|---|---|---|
| SibSp | ||
| 0 | 398 | 210 |
| 1 | 97 | 112 |
| 2 | 15 | 13 |
| 3 | 12 | 4 |
| 4 | 15 | 3 |
| 5 | 5 | 0 |
| 8 | 7 | 0 |
f,ax=plt.subplots(1,2,figsize=(20,8))
sns.barplot('SibSp','Survived', data=train,ax=ax[0])
ax[0].set_title('SipSp vs Survived in BarPlot')
sns.factorplot('SibSp','Survived', data=train,ax=ax[1])
ax[1].set_title('SibSp vs Survived in FactorPlot')
plt.close(2)
plt.show()

pd.crosstab(train.SibSp,train.Pclass).style.background_gradient('summer_r')
| Pclass | 1 | 2 | 3 |
|---|---|---|---|
| SibSp | |||
| 0 | 137 | 120 | 351 |
| 1 | 71 | 55 | 83 |
| 2 | 5 | 8 | 15 |
| 3 | 3 | 1 | 12 |
| 4 | 0 | 0 | 18 |
| 5 | 0 | 0 | 5 |
| 8 | 0 | 0 | 7 |
f,ax=plt.subplots(1,2,figsize=(20,8))
sns.barplot('Pclass','Survived', data=train,ax=ax[0])
ax[0].set_title('Pclass vs Survived in BarPlot')
sns.factorplot('Pclass','Survived', data=train,ax=ax[1])
ax[1].set_title('Pclass vs Survived in FactorPlot')
plt.close(2)
plt.show()

sns.countplot(train['Survived'],label="Count")
<matplotlib.axes._subplots.AxesSubplot at 0x1be652cdc48>

4.7 Embarked
FacetGrid = sns.FacetGrid(train, row='Embarked', size=4.5, aspect=1.6)
FacetGrid.map(sns.pointplot, 'Pclass', 'Survived', 'Sex', palette=None, order=None, hue_order=None )
FacetGrid.add_legend()
<seaborn.axisgrid.FacetGrid at 0x1be65530cc8>

Pclass1 = train[train['Pclass']==1]['Embarked'].value_counts()
Pclass2 = train[train['Pclass']==2]['Embarked'].value_counts()
Pclass3 = train[train['Pclass']==3]['Embarked'].value_counts()
df = pd.DataFrame([Pclass1, Pclass2, Pclass3])
df.index = ['1st class','2nd class', '3rd class']
df.plot(kind='bar',stacked=True, figsize=(10,5))
<matplotlib.axes._subplots.AxesSubplot at 0x1be654a8388>

fill out missing embark with S embark
for dataset in train_test_data:
dataset['Embarked'] = dataset['Embarked'].fillna('S')
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S | 0 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C | 2 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S | 1 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 53.1000 | C123 | S | 2 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 8.0500 | NaN | S | 0 |
embarked_mapping = {"S": 0, "C": 1, "Q": 2}
for dataset in train_test_data:
dataset['Embarked'] = dataset['Embarked'].map(embarked_mapping)
train.Embarked.isnull().any()
False
#Look at survival rate by sex, age and class
age = pd.cut(train['Age'], [0, 18, 80])
train.pivot_table('Survived', ['Sex', age], 'Pclass')
| Pclass | 1 | 2 | 3 | |
|---|---|---|---|---|
| Sex | Age | |||
| 0 | (0, 18] | 0.352941 | 0.082474 | 0.114379 |
| 1 | (0, 18] | 0.977273 | 0.909091 | 0.486486 |
4.8 Fare price
# fill for missing value with the median fare for each class:
train['Fare'].fillna(train.groupby('Pclass')['Fare'].transform('median'), inplace= True)
test['Fare'].fillna(test.groupby('Pclass')['Fare'].transform('median'), inplace= True)
#Plot the Prices Paid Of Each Class
plt.scatter(train['Fare'], train['Pclass'], color = 'purple', label='Passenger Paid')
plt.ylabel('Class')
plt.xlabel('Price / Fare')
plt.title('Price Of Each Class')
plt.legend()
plt.show()

facet =sns.FacetGrid(train, hue='Survived',aspect=4)
facet.map(sns.kdeplot,'Fare',shade=True)
facet.set(xlim=(0, train['Fare'].max()))
facet.add_legend()
plt.show()

facet =sns.FacetGrid(train, hue='Survived',aspect=4)
facet.map(sns.kdeplot,'Fare',shade=True)
facet.set(xlim=(0, train['Fare'].max()))
facet.add_legend()
plt.xlim(0,200)
(0, 200)

for dataset in train_test_data:
dataset.loc[ dataset['Fare'] <= 17, 'Fare'] = 0,
dataset.loc[(dataset['Fare'] > 17) & (dataset['Fare'] <= 30), 'Fare'] = 1,
dataset.loc[(dataset['Fare'] > 30) & (dataset['Fare'] <= 100), 'Fare'] = 2,
dataset.loc[ dataset['Fare'] > 100, 'Fare'] = 3
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 0.0 | NaN | 0 | 0 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 2.0 | C85 | 1 | 2 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 0.0 | NaN | 0 | 1 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 2.0 | C123 | 0 | 2 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 0.0 | NaN | 0 | 0 |
train.Fare.isnull().any()
False
4.9 Cabins
train.Cabin.value_counts()
B96 B98 4
C23 C25 C27 4
G6 4
E101 3
C22 C26 3
..
A26 1
B86 1
B4 1
A10 1
C110 1
Name: Cabin, Length: 147, dtype: int64
for dataset in train_test_data:
dataset['Cabin']=dataset['Cabin'].str[:1]
Pclass1 = train[train['Pclass']==1]['Cabin'].value_counts()
Pclass2 = train[train['Pclass']==2]['Cabin'].value_counts()
Pclass3 = train[train['Pclass']==3]['Cabin'].value_counts()
df=pd.DataFrame([Pclass1, Pclass2, Pclass3])
df.index= ['1st class','2nd class','3rd class']
df.plot(kind = 'bar', stacked = True, figsize =(10,5))
<matplotlib.axes._subplots.AxesSubplot at 0x1be673cf4c8>

cabin_mapping = {"A": 0, "B": 0.4, "C": 0.8, "D": 1.2, "E": 1.6, "F": 2, "G": 2.4, "T": 2.8}
for dataset in train_test_data:
dataset['Cabin'] = dataset['Cabin'].map(cabin_mapping)
# fill missing Fare with median fare for each Pclass
train["Cabin"].fillna(train.groupby("Pclass")["Cabin"].transform("median"), inplace=True)
test["Cabin"].fillna(test.groupby("Pclass")["Cabin"].transform("median"), inplace=True)
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 0.0 | 2.0 | 0 | 0 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 2.0 | 0.8 | 1 | 2 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 0.0 | 2.0 | 0 | 1 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 2.0 | 0.8 | 0 | 2 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 0.0 | 2.0 | 0 | 0 |
train.Embarked.isnull().any()
False
4.10 FamilySize
train["FamilySize"] = train["SibSp"] + train["Parch"] + 1
test["FamilySize"] = test["SibSp"] + test["Parch"] + 1
facet = sns.FacetGrid(train, hue="Survived",aspect=4)
facet.map(sns.kdeplot,'FamilySize',shade= True)
facet.set(xlim=(0, train['FamilySize'].max()))
facet.add_legend()
plt.xlim(0)
(0, 11.0)

family_mapping = {1: 0, 2: 0.4, 3: 0.8, 4: 1.2, 5: 1.6, 6: 2, 7: 2.4, 8: 2.8, 9: 3.2, 10: 3.6, 11: 4}
for dataset in train_test_data:
dataset['FamilySize'] = dataset['FamilySize'].map(family_mapping)
train.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | Title | FamilySize | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1.0 | 1 | 0 | A/5 21171 | 0.0 | 2.0 | 0 | 0 | 0.4 |
| 1 | 2 | 1 | 1 | 1 | 3.0 | 1 | 0 | PC 17599 | 2.0 | 0.8 | 1 | 2 | 0.4 |
| 2 | 3 | 1 | 3 | 1 | 1.0 | 0 | 0 | STON/O2. 3101282 | 0.0 | 2.0 | 0 | 1 | 0.0 |
| 3 | 4 | 1 | 1 | 1 | 2.0 | 1 | 0 | 113803 | 2.0 | 0.8 | 0 | 2 | 0.4 |
| 4 | 5 | 0 | 3 | 0 | 2.0 | 0 | 0 | 373450 | 0.0 | 2.0 | 0 | 0 | 0.0 |
#Print the unique values in the columns
print(train['Sex'].unique())
print(train['Embarked'].unique())
['male' 'female']
['S' 'C' 'Q' nan]
5. Modeling
5.1 Supervised Learning
# Importing Classifier Modules
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
import numpy as np
train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 13 columns):
PassengerId 891 non-null int64
Survived 891 non-null int64
Pclass 891 non-null int64
Sex 891 non-null int64
Age 891 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Ticket 891 non-null object
Fare 891 non-null float64
Cabin 891 non-null float64
Embarked 891 non-null int64
Title 891 non-null int64
FamilySize 891 non-null float64
dtypes: float64(4), int64(8), object(1)
memory usage: 90.6+ KB
features_drop = ['Ticket', 'SibSp', 'Parch']
train = train.drop(features_drop, axis=1)
test = test.drop(features_drop, axis=1)
train = train.drop(['PassengerId'], axis=1)
train_data = train.drop('Survived', axis=1)
target = train['Survived']
train_data.shape, target.shape
((891, 8), (891,))
train_data.head()
| Pclass | Sex | Age | Fare | Cabin | Embarked | Title | FamilySize | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 0 | 1.0 | 0.0 | 2.0 | 0 | 0 | 0.4 |
| 1 | 1 | 1 | 3.0 | 2.0 | 0.8 | 1 | 2 | 0.4 |
| 2 | 3 | 1 | 1.0 | 0.0 | 2.0 | 0 | 1 | 0.0 |
| 3 | 1 | 1 | 2.0 | 2.0 | 0.8 | 0 | 2 | 0.4 |
| 4 | 3 | 0 | 2.0 | 0.0 | 2.0 | 0 | 0 | 0.0 |
target.head()
0 0
1 1
2 1
3 1
4 0
Name: Survived, dtype: int64
colormap = plt.cm.viridis
plt.figure(figsize=(12,12))
plt.title('Pearson Correlation of Features', y=1.05, size=15)
sns.heatmap(train.astype(float).corr(),linewidths=0.1,vmax=1.0, square=True, cmap=colormap, linecolor='white', annot=True)
<matplotlib.axes._subplots.AxesSubplot at 0x1be6750f088>

# Split the dataset into 80% Training set and 20% Testing set
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(train_data, target, test_size = 0.2, random_state = 0)
# Scale the data to bring all features to the same level of magnitude
# This means the data will be within a specific range for example 0 -100 or 0 - 1
#Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
def models(X_train,Y_train):
#Using Logistic Regression Algorithm to the Training Set
from sklearn.linear_model import LogisticRegression
log = LogisticRegression(random_state = 0)
log.fit(X_train, Y_train)
#Using KNeighborsClassifier Method of neighbors class to use Nearest Neighbor algorithm
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors = 5, metric = 'minkowski', p = 2)
knn.fit(X_train, Y_train)
#Using SVC method of svm class to use Support Vector Machine Algorithm
from sklearn.svm import SVC
svc_lin = SVC(kernel = 'linear', random_state = 0)
svc_lin.fit(X_train, Y_train)
#Using SVC method of svm class to use Kernel SVM Algorithm
from sklearn.svm import SVC
svc_rbf = SVC(kernel = 'rbf', random_state = 0)
svc_rbf.fit(X_train, Y_train)
#Using GaussianNB method of naïve_bayes class to use Naïve Bayes Algorithm
from sklearn.naive_bayes import GaussianNB
gauss = GaussianNB()
gauss.fit(X_train, Y_train)
#Using DecisionTreeClassifier of tree class to use Decision Tree Algorithm
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(criterion = 'entropy', random_state = 0)
tree.fit(X_train, Y_train)
#Using RandomForestClassifier method of ensemble class to use Random Forest Classification algorithm
from sklearn.ensemble import RandomForestClassifier
forest = RandomForestClassifier(n_estimators = 10, criterion = 'entropy', random_state = 0)
forest.fit(X_train, Y_train)
#print model accuracy on the training data.
print('[0]Logistic Regression Training Accuracy:', log.score(X_train, Y_train))
print('[1]K Nearest Neighbor Training Accuracy:', knn.score(X_train, Y_train))
print('[2]Support Vector Machine (Linear Classifier) Training Accuracy:', svc_lin.score(X_train, Y_train))
print('[3]Support Vector Machine (RBF Classifier) Training Accuracy:', svc_rbf.score(X_train, Y_train))
print('[4]Gaussian Naive Bayes Training Accuracy:', gauss.score(X_train, Y_train))
print('[5]Decision Tree Classifier Training Accuracy:', tree.score(X_train, Y_train))
print('[6]Random Forest Classifier Training Accuracy:', forest.score(X_train, Y_train))
return log, knn, svc_lin, svc_rbf, gauss, tree, forest
#Get and train all of the models
model = models(X_train,Y_train)
[0]Logistic Regression Training Accuracy: 0.8314606741573034
[1]K Nearest Neighbor Training Accuracy: 0.8553370786516854
[2]Support Vector Machine (Linear Classifier) Training Accuracy: 0.8286516853932584
[3]Support Vector Machine (RBF Classifier) Training Accuracy: 0.848314606741573
[4]Gaussian Naive Bayes Training Accuracy: 0.7823033707865169
[5]Decision Tree Classifier Training Accuracy: 0.901685393258427
[6]Random Forest Classifier Training Accuracy: 0.8932584269662921
#Show the confusion matrix and accuracy for all of the models on the test data
#Classification accuracy is the ratio of correct predictions to total predictions made.
from sklearn.metrics import confusion_matrix
for i in range(len(model)):
cm = confusion_matrix(Y_test, model[i].predict(X_test))
#extracting true_positives, false_positives, true_negatives, false_negatives
TN, FP, FN, TP = confusion_matrix(Y_test, model[i].predict(X_test)).ravel()
print(cm)
print('Model[{}] Testing Accuracy = "{} !"'.format(i, (TP + TN) / (TP + TN + FN + FP)))
print()# Print a new line
[[92 18]
[17 52]]
Model[0] Testing Accuracy = "0.8044692737430168 !"
[[97 13]
[20 49]]
Model[1] Testing Accuracy = "0.8156424581005587 !"
[[91 19]
[17 52]]
Model[2] Testing Accuracy = "0.7988826815642458 !"
[[100 10]
[ 22 47]]
Model[3] Testing Accuracy = "0.8212290502793296 !"
[[82 28]
[ 7 62]]
Model[4] Testing Accuracy = "0.8044692737430168 !"
[[97 13]
[25 44]]
Model[5] Testing Accuracy = "0.7877094972067039 !"
[[100 10]
[ 23 46]]
Model[6] Testing Accuracy = "0.8156424581005587 !"
#Get Feature importance
forest=model[6]
importances = pd.DataFrame({'feature':train_data.iloc[:,0:8].columns, 'importance': np.round(forest.feature_importances_, 3)})
importances= importances.sort_values('importance', ascending=False).set_index('feature')
importances
| importance | |
|---|---|
| feature | |
| Title | 0.257 |
| Sex | 0.135 |
| FamilySize | 0.129 |
| Age | 0.127 |
| Cabin | 0.107 |
| Fare | 0.097 |
| Pclass | 0.084 |
| Embarked | 0.064 |
#Visualize the importance
importances.plot.bar()
<matplotlib.axes._subplots.AxesSubplot at 0x1be649b7b48>

#print the prediction of the random forest classifier
pred = model[6].predict(X_test)
print(pred)
print()
#print the actual value
print(Y_test)
[0 0 0 1 0 0 1 1 0 1 0 1 0 1 1 1 0 0 0 1 0 1 0 0 1 1 0 1 1 0 0 1 0 0 0 0 0
0 0 0 0 0 0 0 1 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 1 0 0 0
0 1 0 0 0 1 0 1 1 0 0 1 1 0 1 0 0 0 1 1 0 0 1 0 1 0 0 0 0 0 0 1 1 0 0 1 0
1 0 1 0 1 1 1 0 1 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 0 1 1 1 0 1
1 0 0 0 0 0 1 0 0 0 1 1 0 0 1 0 0 0 0 0 0 0 0 1 0 0 1 0 1 0 0]
495 0
648 0
278 0
31 1
255 1
..
780 1
837 0
215 1
833 0
372 0
Name: Survived, Length: 179, dtype: int64
#my survival
#Pclass Sex Age Fare Cabin Embarked Title FamilySize
#scaling my survival
my_survival = [[2, 0, 3, 2, 0.8, 0, 0, 0.8]]
#scaling:
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
my_survival_scaled = sc.fit_transform(my_survival)
#print prediction of my survival using Random forest calssifier:
pred= model[6].predict(my_survival_scaled)
print(pred)
if pred == 0:
print('Oh no you didn not survive')
else:
print('nice! you survived!')
[1]
nice! you survived!
ROC and AUC using logistic regression as an example
#Using Logistic Regression Algorithm to the Training Set
from sklearn.linear_model import LogisticRegression
log = LogisticRegression(random_state = 0)
log.fit(X_train, Y_train)
Y_predict=log.predict(X_test)
from sklearn.metrics import accuracy_score
print(accuracy_score(Y_test,Y_predict))
pd.crosstab(Y_test, Y_predict)
0.8044692737430168
| col_0 | 0 | 1 |
|---|---|---|
| Survived | ||
| 0 | 92 | 18 |
| 1 | 17 | 52 |
log.predict_proba(X_test)
array([[0.90562449, 0.09437551],
[0.92146315, 0.07853685],
[0.66984458, 0.33015542],
[0.03898146, 0.96101854],
[0.39961516, 0.60038484],
[0.56001427, 0.43998573],
[0.09451128, 0.90548872],
[0.13543067, 0.86456933],
[0.52243664, 0.47756336],
[0.21745097, 0.78254903],
[0.91920705, 0.08079295],
[0.31239561, 0.68760439],
[0.8869834 , 0.1130166 ],
[0.176777 , 0.823223 ],
[0.04009628, 0.95990372],
[0.22778632, 0.77221368],
[0.88025683, 0.11974317],
[0.81108283, 0.18891717],
[0.92146315, 0.07853685],
[0.3501963 , 0.6498037 ],
[0.73578675, 0.26421325],
[0.03936127, 0.96063873],
[0.8869834 , 0.1130166 ],
[0.56001427, 0.43998573],
[0.32009766, 0.67990234],
[0.07297558, 0.92702442],
[0.92146315, 0.07853685],
[0.32009766, 0.67990234],
[0.09101505, 0.90898495],
[0.67756724, 0.32243276],
[0.90562449, 0.09437551],
[0.31239561, 0.68760439],
[0.92146315, 0.07853685],
[0.56001427, 0.43998573],
[0.9478045 , 0.0521955 ],
[0.53010852, 0.46989148],
[0.94930607, 0.05069393],
[0.8163354 , 0.1836646 ],
[0.73578675, 0.26421325],
[0.87645115, 0.12354885],
[0.81178102, 0.18821898],
[0.85739412, 0.14260588],
[0.92146315, 0.07853685],
[0.96340222, 0.03659778],
[0.05396473, 0.94603527],
[0.92146315, 0.07853685],
[0.92146315, 0.07853685],
[0.17244067, 0.82755933],
[0.87645115, 0.12354885],
[0.80556426, 0.19443574],
[0.5941448 , 0.4058552 ],
[0.48225335, 0.51774665],
[0.176777 , 0.823223 ],
[0.88025683, 0.11974317],
[0.56475591, 0.43524409],
[0.81108283, 0.18891717],
[0.71688627, 0.28311373],
[0.72293216, 0.27706784],
[0.75205183, 0.24794817],
[0.93781784, 0.06218216],
[0.85739412, 0.14260588],
[0.60996825, 0.39003175],
[0.13191367, 0.86808633],
[0.51004073, 0.48995927],
[0.44867709, 0.55132291],
[0.8869834 , 0.1130166 ],
[0.24865623, 0.75134377],
[0.15615491, 0.84384509],
[0.176777 , 0.823223 ],
[0.06453911, 0.93546089],
[0.09101505, 0.90898495],
[0.75868937, 0.24131063],
[0.61702161, 0.38297839],
[0.92146315, 0.07853685],
[0.88025683, 0.11974317],
[0.27682631, 0.72317369],
[0.85032561, 0.14967439],
[0.67436994, 0.32563006],
[0.92146315, 0.07853685],
[0.78425929, 0.21574071],
[0.90693144, 0.09306856],
[0.50490792, 0.49509208],
[0.13417399, 0.86582601],
[0.82161595, 0.17838405],
[0.8505506 , 0.1494494 ],
[0.04500599, 0.95499401],
[0.07865182, 0.92134818],
[0.46394494, 0.53605506],
[0.11003162, 0.88996838],
[0.44746386, 0.55253614],
[0.68184598, 0.31815402],
[0.88025683, 0.11974317],
[0.22942292, 0.77057708],
[0.0746709 , 0.9253291 ],
[0.67756724, 0.32243276],
[0.90562449, 0.09437551],
[0.16189054, 0.83810946],
[0.96340222, 0.03659778],
[0.09570488, 0.90429512],
[0.48994053, 0.51005947],
[0.98438827, 0.01561173],
[0.26908461, 0.73091539],
[0.88025683, 0.11974317],
[0.94626097, 0.05373903],
[0.42887836, 0.57112164],
[0.39500171, 0.60499829],
[0.11403093, 0.88596907],
[0.75605068, 0.24394932],
[0.77567238, 0.22432762],
[0.32768611, 0.67231389],
[0.94930607, 0.05069393],
[0.06305814, 0.93694186],
[0.88306269, 0.11693731],
[0.32009766, 0.67990234],
[0.80401578, 0.19598422],
[0.22158155, 0.77841845],
[0.2478141 , 0.7521859 ],
[0.02291349, 0.97708651],
[0.94930607, 0.05069393],
[0.283029 , 0.716971 ],
[0.88306269, 0.11693731],
[0.8869834 , 0.1130166 ],
[0.88025683, 0.11974317],
[0.11654445, 0.88345555],
[0.92146315, 0.07853685],
[0.69177453, 0.30822547],
[0.90562449, 0.09437551],
[0.91920705, 0.08079295],
[0.8163354 , 0.1836646 ],
[0.22869217, 0.77130783],
[0.23949934, 0.76050066],
[0.88025683, 0.11974317],
[0.88025683, 0.11974317],
[0.42151289, 0.57848711],
[0.80164361, 0.19835639],
[0.88025683, 0.11974317],
[0.92393466, 0.07606534],
[0.45630075, 0.54369925],
[0.8980445 , 0.1019555 ],
[0.73578675, 0.26421325],
[0.8163354 , 0.1836646 ],
[0.07782203, 0.92217797],
[0.92146315, 0.07853685],
[0.23949934, 0.76050066],
[0.10297104, 0.89702896],
[0.32009766, 0.67990234],
[0.8163354 , 0.1836646 ],
[0.20322749, 0.79677251],
[0.06080351, 0.93919649],
[0.92146315, 0.07853685],
[0.75868937, 0.24131063],
[0.42903417, 0.57096583],
[0.54530973, 0.45469027],
[0.92146315, 0.07853685],
[0.12729888, 0.87270112],
[0.73578675, 0.26421325],
[0.40702062, 0.59297938],
[0.87645115, 0.12354885],
[0.23949934, 0.76050066],
[0.4177658 , 0.5822342 ],
[0.92146315, 0.07853685],
[0.85739412, 0.14260588],
[0.06930139, 0.93069861],
[0.5574304 , 0.4425696 ],
[0.90023128, 0.09976872],
[0.88025683, 0.11974317],
[0.94930607, 0.05069393],
[0.88025683, 0.11974317],
[0.88025683, 0.11974317],
[0.88025683, 0.11974317],
[0.94930607, 0.05069393],
[0.06930139, 0.93069861],
[0.92146315, 0.07853685],
[0.92146315, 0.07853685],
[0.19436312, 0.80563688],
[0.92146315, 0.07853685],
[0.07096286, 0.92903714],
[0.88025683, 0.11974317],
[0.88025683, 0.11974317]])
log.predict_proba(X_test)[:,1]
array([0.09437551, 0.07853685, 0.33015542, 0.96101854, 0.60038484,
0.43998573, 0.90548872, 0.86456933, 0.47756336, 0.78254903,
0.08079295, 0.68760439, 0.1130166 , 0.823223 , 0.95990372,
0.77221368, 0.11974317, 0.18891717, 0.07853685, 0.6498037 ,
0.26421325, 0.96063873, 0.1130166 , 0.43998573, 0.67990234,
0.92702442, 0.07853685, 0.67990234, 0.90898495, 0.32243276,
0.09437551, 0.68760439, 0.07853685, 0.43998573, 0.0521955 ,
0.46989148, 0.05069393, 0.1836646 , 0.26421325, 0.12354885,
0.18821898, 0.14260588, 0.07853685, 0.03659778, 0.94603527,
0.07853685, 0.07853685, 0.82755933, 0.12354885, 0.19443574,
0.4058552 , 0.51774665, 0.823223 , 0.11974317, 0.43524409,
0.18891717, 0.28311373, 0.27706784, 0.24794817, 0.06218216,
0.14260588, 0.39003175, 0.86808633, 0.48995927, 0.55132291,
0.1130166 , 0.75134377, 0.84384509, 0.823223 , 0.93546089,
0.90898495, 0.24131063, 0.38297839, 0.07853685, 0.11974317,
0.72317369, 0.14967439, 0.32563006, 0.07853685, 0.21574071,
0.09306856, 0.49509208, 0.86582601, 0.17838405, 0.1494494 ,
0.95499401, 0.92134818, 0.53605506, 0.88996838, 0.55253614,
0.31815402, 0.11974317, 0.77057708, 0.9253291 , 0.32243276,
0.09437551, 0.83810946, 0.03659778, 0.90429512, 0.51005947,
0.01561173, 0.73091539, 0.11974317, 0.05373903, 0.57112164,
0.60499829, 0.88596907, 0.24394932, 0.22432762, 0.67231389,
0.05069393, 0.93694186, 0.11693731, 0.67990234, 0.19598422,
0.77841845, 0.7521859 , 0.97708651, 0.05069393, 0.716971 ,
0.11693731, 0.1130166 , 0.11974317, 0.88345555, 0.07853685,
0.30822547, 0.09437551, 0.08079295, 0.1836646 , 0.77130783,
0.76050066, 0.11974317, 0.11974317, 0.57848711, 0.19835639,
0.11974317, 0.07606534, 0.54369925, 0.1019555 , 0.26421325,
0.1836646 , 0.92217797, 0.07853685, 0.76050066, 0.89702896,
0.67990234, 0.1836646 , 0.79677251, 0.93919649, 0.07853685,
0.24131063, 0.57096583, 0.45469027, 0.07853685, 0.87270112,
0.26421325, 0.59297938, 0.12354885, 0.76050066, 0.5822342 ,
0.07853685, 0.14260588, 0.93069861, 0.4425696 , 0.09976872,
0.11974317, 0.05069393, 0.11974317, 0.11974317, 0.11974317,
0.05069393, 0.93069861, 0.07853685, 0.07853685, 0.80563688,
0.07853685, 0.92903714, 0.11974317, 0.11974317])
np.where(log.predict_proba(X_test)[:,1] >0.3,1,0) #threshold 0.3
array([0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 0, 0, 0, 1, 0, 1,
0, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,
1, 0, 0, 1, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0,
1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 1,
1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1,
0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 1, 0,
0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 0,
1, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0,
1, 0, 0])
th3= np.where(log.predict_proba(X_test)[:,1] >0.3,1,0) #threshold 0.3
th4=np.where(log.predict_proba(X_test)[:,1] >0.4,1,0) #threshold 0.4
th5=np.where(log.predict_proba(X_test)[:,1] >0.5,1,0) #threshold 0.5
th6=np.where(log.predict_proba(X_test)[:,1] >0.6,1,0) #threshold 0.6
th8=np.where(log.predict_proba(X_test)[:,1] >0.8,1,0) #threshold 0.6
pd.crosstab(Y_test, th3)
| col_0 | 0 | 1 |
|---|---|---|
| Survived | ||
| 0 | 81 | 29 |
| 1 | 9 | 60 |
pd.crosstab(Y_test, th4)
| col_0 | 0 | 1 |
|---|---|---|
| Survived | ||
| 0 | 89 | 21 |
| 1 | 9 | 60 |
pd.crosstab(Y_test, th5)
| col_0 | 0 | 1 |
|---|---|---|
| Survived | ||
| 0 | 92 | 18 |
| 1 | 17 | 52 |
pd.crosstab(Y_test, th6)
| col_0 | 0 | 1 |
|---|---|---|
| Survived | ||
| 0 | 97 | 13 |
| 1 | 23 | 46 |
def predict_thrs (log,X_test,thrs):
import numpy as np
Y_predict = np.where(log.predict_proba(X_test)[:,1] >thrs,1,0)
return Y_predict
import numpy as np
from sklearn.metrics import confusion_matrix
for thr in np.arange(0,1.1,0.1):
Y_predict= predict_thrs(log, X_test,thr)
print("Threshold :", thr)
print(confusion_matrix(Y_test, Y_predict))
Threshold : 0.0
[[ 0 110]
[ 0 69]]
Threshold : 0.1
[[35 75]
[ 2 67]]
Threshold : 0.2
[[70 40]
[ 8 61]]
Threshold : 0.30000000000000004
[[81 29]
[ 9 60]]
Threshold : 0.4
[[89 21]
[ 9 60]]
Threshold : 0.5
[[92 18]
[17 52]]
Threshold : 0.6000000000000001
[[97 13]
[23 46]]
Threshold : 0.7000000000000001
[[100 10]
[ 30 39]]
Threshold : 0.8
[[106 4]
[ 38 31]]
Threshold : 0.9
[[109 1]
[ 50 19]]
Threshold : 1.0
[[110 0]
[ 69 0]]
from sklearn.metrics import roc_curve, roc_auc_score
tpr,fpr,thrs= roc_curve(Y_test, log.predict_proba(X_test)[:,1])
tpr
array([0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0.00909091, 0.00909091, 0.01818182,
0.01818182, 0.02727273, 0.02727273, 0.03636364, 0.03636364,
0.03636364, 0.03636364, 0.05454545, 0.05454545, 0.08181818,
0.08181818, 0.09090909, 0.09090909, 0.09090909, 0.11818182,
0.11818182, 0.12727273, 0.12727273, 0.14545455, 0.14545455,
0.15454545, 0.15454545, 0.16363636, 0.16363636, 0.17272727,
0.17272727, 0.18181818, 0.18181818, 0.19090909, 0.19090909,
0.22727273, 0.24545455, 0.28181818, 0.31818182, 0.33636364,
0.34545455, 0.37272727, 0.37272727, 0.38181818, 0.4 ,
0.4 , 0.43636364, 0.46363636, 0.5 , 0.63636364,
0.63636364, 0.67272727, 0.69090909, 0.72727273, 0.73636364,
0.74545455, 0.9 , 0.90909091, 0.90909091, 0.92727273,
0.97272727, 0.99090909, 1. ])
fpr
array([0. , 0.01449275, 0.13043478, 0.15942029, 0.23188406,
0.26086957, 0.27536232, 0.27536232, 0.31884058, 0.31884058,
0.34782609, 0.34782609, 0.36231884, 0.36231884, 0.39130435,
0.43478261, 0.49275362, 0.49275362, 0.50724638, 0.50724638,
0.53623188, 0.53623188, 0.56521739, 0.5942029 , 0.60869565,
0.66666667, 0.66666667, 0.68115942, 0.68115942, 0.69565217,
0.69565217, 0.71014493, 0.71014493, 0.76811594, 0.76811594,
0.79710145, 0.79710145, 0.8115942 , 0.84057971, 0.86956522,
0.86956522, 0.86956522, 0.86956522, 0.86956522, 0.86956522,
0.88405797, 0.88405797, 0.89855072, 0.89855072, 0.89855072,
0.91304348, 0.91304348, 0.91304348, 0.94202899, 0.94202899,
0.97101449, 0.97101449, 0.97101449, 0.97101449, 0.97101449,
0.98550725, 0.98550725, 0.98550725, 1. , 1. ,
1. , 1. , 1. ])
thrs
array([1.97708651, 0.97708651, 0.93546089, 0.93069861, 0.92134818,
0.90898495, 0.90548872, 0.90429512, 0.88596907, 0.88345555,
0.86808633, 0.86582601, 0.86456933, 0.84384509, 0.82755933,
0.823223 , 0.77841845, 0.77130783, 0.77057708, 0.76050066,
0.75134377, 0.73091539, 0.716971 , 0.68760439, 0.67990234,
0.60038484, 0.59297938, 0.5822342 , 0.57112164, 0.57096583,
0.55253614, 0.55132291, 0.54369925, 0.49509208, 0.48995927,
0.46989148, 0.45469027, 0.4425696 , 0.43998573, 0.4058552 ,
0.32563006, 0.32243276, 0.27706784, 0.26421325, 0.24394932,
0.24131063, 0.19835639, 0.19598422, 0.19443574, 0.18891717,
0.18821898, 0.1836646 , 0.1494494 , 0.12354885, 0.11974317,
0.11693731, 0.1130166 , 0.09976872, 0.09437551, 0.09306856,
0.08079295, 0.07853685, 0.07606534, 0.06218216, 0.0521955 ,
0.05069393, 0.03659778, 0.01561173])
import matplotlib.pyplot as plt
from sklearn.metrics import roc_curve,roc_auc_score
%matplotlib inline
fpr,tpr,thrs= roc_curve(Y_test, log.predict_proba(X_test)[:,1])
plt.xlim([0.0,1.0])
plt.ylim([0.0,1.0])
plt.plot(fpr,tpr,color='red')
plt.show()

roc_auc_score(Y_test, log.predict_proba(X_test)[:,1])
0.8704216073781291
5.2 Unsupervised learning:
5.2.1 Kmeans
from sklearn.cluster import KMeans
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from matplotlib import pyplot as plt
%matplotlib inline
df = pd.read_csv('Unsupervised.csv')
df.head()
| Name | Age | Fare | |
|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 22.0 | 7.2500 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 | 71.2833 |
| 2 | Heikkinen, Miss. Laina | 26.0 | 7.9250 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 | 53.1000 |
| 4 | Allen, Mr. William Henry | 35.0 | 8.0500 |
df.isnull().sum()
Name 0
Age 177
Fare 0
dtype: int64
df['Initial']=0
for i in df:
df['Initial']=df.Name.str.extract('([A-Za-z]+)\.') #extracting Name initials
pd.crosstab(df.Initial,train_data.Sex).T.style.background_gradient(cmap='summer_r')
| Initial | Capt | Col | Countess | Don | Dr | Jonkheer | Lady | Major | Master | Miss | Mlle | Mme | Mr | Mrs | Ms | Rev | Sir |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||||||||||
| 0 | 1 | 2 | 0 | 1 | 6 | 1 | 0 | 2 | 40 | 0 | 0 | 0 | 517 | 0 | 0 | 6 | 1 |
| 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 182 | 2 | 1 | 0 | 125 | 1 | 0 | 0 |
df['Initial'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess',
'Jonkheer','Col','Rev','Capt','Sir','Don'],['Miss',
'Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr'],inplace=True)
df.groupby('Initial')['Age'].mean()
Initial
Master 4.574167
Miss 21.860000
Mr 32.739609
Mrs 35.981818
Other 45.888889
Name: Age, dtype: float64
df.loc[(df.Age.isnull()) & (df.Initial=='Mr'),'Age']=33
df.loc[(df.Age.isnull()) & (df.Initial=='Mrs'),'Age']=36
df.loc[(df.Age.isnull()) & (df.Initial=='Master'),'Age']=5
df.loc[(df.Age.isnull()) & (df.Initial=='Miss'),'Age']=22
df.loc[(df.Age.isnull()) & (df.Initial=='Other'),'Age']=46
df.Age.isnull().any()
False
plt.scatter(df.Age,df.Fare)
<matplotlib.collections.PathCollection at 0x1be6786f688>

km = KMeans(n_clusters=3)
km
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
y_predicted= km.fit_predict(df[['Age','Fare']])
y_predicted
array([1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 2, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 1,
1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1,
0, 1, 1, 1, 1, 1, 1, 1, 2, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1,
1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1,
1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1,
1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 0, 2, 1, 1, 1, 0, 1,
1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 0, 1, 2, 1, 1, 1, 1, 1, 0, 0, 0,
1, 0, 0, 2, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 1, 1, 0,
1, 1, 0, 1, 0, 1, 0, 0, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 0,
1, 0, 1, 2, 1, 1, 2, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 2, 1,
1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1,
0, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0,
1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 2,
1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1,
0, 1, 1, 1, 1, 1, 1, 2, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0,
1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1,
1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0,
1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0,
0, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 2, 1, 0,
1, 1, 1, 1, 1, 1, 1, 2, 0, 1, 0, 1, 1, 1, 1, 1, 0, 1, 2, 1, 1, 1,
1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 0, 1,
1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 0, 2, 1, 1, 0, 1, 1,
0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1,
0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 0,
1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1,
1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1])
df['cluster']=y_predicted
df.head()
| Name | Age | Fare | Initial | cluster | |
|---|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 22.0 | 7.2500 | Mr | 1 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 | 71.2833 | Mrs | 0 |
| 2 | Heikkinen, Miss. Laina | 26.0 | 7.9250 | Miss | 1 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 | 53.1000 | Mrs | 0 |
| 4 | Allen, Mr. William Henry | 35.0 | 8.0500 | Mr | 1 |
%pylab inline
Populating the interactive namespace from numpy and matplotlib
df1=df[df.cluster==0]
df2=df[df.cluster==1]
df3=df[df.cluster==2]
plt.scatter(df1.Age,df1['Fare'],color = 'green')
plt.scatter(df2.Age,df2['Fare'],color = 'red')
plt.scatter(df3.Age,df3['Fare'],color = 'black')
plt.xlabel('Age')
plt.ylabel('Fare')
plt.legend()
No handles with labels found to put in legend.
<matplotlib.legend.Legend at 0x1be67b69188>

df1=df[df.cluster==0]
df2=df[df.cluster==1]
df3=df[df.cluster==2]
plt.scatter(df1['Fare'],df1.Age,color = 'green')
plt.scatter(df2['Fare'],df2.Age,color = 'red')
plt.scatter(df3['Fare'],df3.Age,color = 'black')
plt.ylabel('Age')
plt.xlabel('Fare')
plt.legend()
No handles with labels found to put in legend.
<matplotlib.legend.Legend at 0x1be677f7fc8>

df
| Name | Age | Fare | Initial | cluster | |
|---|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 22.0 | 7.2500 | Mr | 2 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 | 71.2833 | Mrs | 0 |
| 2 | Heikkinen, Miss. Laina | 26.0 | 7.9250 | Miss | 2 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 | 53.1000 | Mrs | 0 |
| 4 | Allen, Mr. William Henry | 35.0 | 8.0500 | Mr | 2 |
| ... | ... | ... | ... | ... | ... |
| 886 | Montvila, Rev. Juozas | 27.0 | 13.0000 | Other | 2 |
| 887 | Graham, Miss. Margaret Edith | 19.0 | 30.0000 | Miss | 2 |
| 888 | Johnston, Miss. Catherine Helen "Carrie" | 22.0 | 23.4500 | Miss | 2 |
| 889 | Behr, Mr. Karl Howell | 26.0 | 30.0000 | Mr | 2 |
| 890 | Dooley, Mr. Patrick | 32.0 | 7.7500 | Mr | 2 |
891 rows × 5 columns
sc=MinMaxScaler()
sc.fit(df[['Age']])
df['Age']=sc.transform(df[['Age']])
df
| Name | Age | Fare | Initial | cluster | |
|---|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 0.271174 | 7.2500 | Mr | 1 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 0.472229 | 71.2833 | Mrs | 0 |
| 2 | Heikkinen, Miss. Laina | 0.321438 | 7.9250 | Miss | 1 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 0.434531 | 53.1000 | Mrs | 0 |
| 4 | Allen, Mr. William Henry | 0.434531 | 8.0500 | Mr | 1 |
| ... | ... | ... | ... | ... | ... |
| 886 | Montvila, Rev. Juozas | 0.334004 | 13.0000 | Other | 1 |
| 887 | Graham, Miss. Margaret Edith | 0.233476 | 30.0000 | Miss | 1 |
| 888 | Johnston, Miss. Catherine Helen "Carrie" | 0.271174 | 23.4500 | Miss | 1 |
| 889 | Behr, Mr. Karl Howell | 0.321438 | 30.0000 | Mr | 1 |
| 890 | Dooley, Mr. Patrick | 0.396833 | 7.7500 | Mr | 1 |
891 rows × 5 columns
sc=MinMaxScaler()
sc.fit(df[['Fare']])
df['Fare']=sc.transform(df[['Fare']])
df
| Name | Age | Fare | Initial | cluster | |
|---|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 0.271174 | 0.014151 | Mr | 1 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 0.472229 | 0.139136 | Mrs | 0 |
| 2 | Heikkinen, Miss. Laina | 0.321438 | 0.015469 | Miss | 1 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 0.434531 | 0.103644 | Mrs | 0 |
| 4 | Allen, Mr. William Henry | 0.434531 | 0.015713 | Mr | 1 |
| ... | ... | ... | ... | ... | ... |
| 886 | Montvila, Rev. Juozas | 0.334004 | 0.025374 | Other | 1 |
| 887 | Graham, Miss. Margaret Edith | 0.233476 | 0.058556 | Miss | 1 |
| 888 | Johnston, Miss. Catherine Helen "Carrie" | 0.271174 | 0.045771 | Miss | 1 |
| 889 | Behr, Mr. Karl Howell | 0.321438 | 0.058556 | Mr | 1 |
| 890 | Dooley, Mr. Patrick | 0.396833 | 0.015127 | Mr | 1 |
891 rows × 5 columns
km= KMeans(n_clusters=3)
y_predicted = km.fit_predict(df[['Age', 'Fare']])
y_predicted
array([2, 0, 0, 0, 0, 0, 1, 2, 0, 2, 2, 1, 2, 0, 2, 1, 2, 0, 0, 0, 0, 0,
2, 0, 2, 0, 0, 2, 2, 0, 0, 0, 2, 1, 0, 1, 0, 2, 2, 2, 0, 0, 0, 2,
2, 0, 0, 2, 0, 2, 2, 2, 1, 0, 1, 0, 2, 0, 2, 2, 2, 0, 1, 2, 0, 2,
0, 2, 2, 0, 0, 2, 2, 0, 0, 0, 0, 0, 2, 0, 2, 0, 2, 0, 2, 0, 2, 0,
2, 2, 0, 2, 1, 0, 1, 0, 1, 2, 0, 0, 0, 0, 2, 0, 0, 0, 2, 0, 0, 2,
1, 2, 2, 2, 2, 2, 1, 0, 2, 2, 2, 0, 0, 0, 1, 2, 0, 2, 2, 1, 0, 2,
1, 0, 0, 2, 2, 0, 2, 2, 0, 2, 2, 2, 2, 2, 0, 2, 0, 0, 1, 2, 1, 0,
0, 1, 2, 0, 0, 2, 1, 0, 0, 2, 2, 2, 0, 1, 0, 0, 1, 2, 2, 2, 1, 2,
2, 1, 0, 0, 2, 0, 2, 2, 2, 0, 0, 1, 0, 0, 0, 2, 2, 2, 1, 1, 0, 0,
2, 2, 0, 0, 0, 1, 2, 2, 0, 0, 2, 0, 2, 0, 2, 0, 0, 0, 0, 0, 0, 0,
2, 0, 1, 0, 0, 2, 2, 2, 2, 2, 0, 0, 1, 2, 2, 2, 1, 2, 2, 0, 2, 2,
0, 2, 0, 1, 0, 2, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 2, 1, 0,
2, 0, 2, 0, 1, 0, 0, 0, 0, 0, 2, 1, 1, 0, 2, 0, 1, 0, 2, 2, 0, 0,
0, 2, 0, 2, 0, 2, 0, 2, 2, 0, 2, 2, 0, 1, 2, 0, 2, 2, 0, 2, 2, 2,
0, 0, 2, 2, 0, 0, 1, 0, 2, 1, 0, 1, 2, 0, 0, 2, 0, 0, 1, 0, 0, 2,
2, 1, 1, 2, 0, 0, 0, 1, 1, 1, 2, 2, 0, 0, 0, 2, 0, 0, 2, 0, 2, 0,
2, 0, 0, 0, 2, 0, 2, 2, 0, 0, 1, 0, 0, 0, 1, 0, 2, 2, 0, 2, 2, 2,
2, 0, 2, 0, 2, 2, 1, 2, 0, 0, 0, 2, 2, 0, 0, 2, 0, 2, 0, 2, 2, 2,
0, 1, 2, 0, 0, 0, 2, 0, 2, 0, 1, 2, 2, 2, 0, 0, 0, 0, 1, 0, 0, 2,
0, 2, 0, 2, 0, 0, 2, 0, 0, 2, 0, 0, 0, 0, 0, 2, 1, 2, 2, 2, 1, 0,
1, 2, 0, 0, 0, 2, 2, 0, 2, 1, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 1, 0,
1, 1, 0, 0, 0, 1, 0, 2, 0, 0, 0, 2, 2, 0, 0, 0, 2, 2, 2, 0, 1, 1,
2, 2, 0, 1, 0, 2, 0, 2, 1, 1, 2, 0, 1, 0, 2, 2, 2, 2, 2, 0, 2, 2,
0, 0, 0, 0, 0, 0, 0, 1, 2, 1, 0, 0, 0, 0, 0, 2, 0, 1, 0, 0, 1, 0,
0, 2, 2, 0, 2, 0, 0, 2, 1, 0, 0, 2, 0, 2, 2, 0, 1, 1, 2, 0, 0, 2,
2, 0, 0, 2, 2, 1, 1, 0, 0, 0, 0, 0, 0, 0, 2, 2, 2, 0, 0, 0, 1, 1,
0, 2, 2, 2, 0, 0, 0, 0, 0, 0, 1, 0, 0, 2, 1, 1, 2, 0, 0, 1, 1, 2,
0, 0, 2, 1, 0, 1, 2, 0, 0, 1, 0, 0, 0, 0, 2, 1, 0, 0, 2, 0, 0, 2,
0, 0, 2, 0, 0, 1, 2, 2, 2, 1, 1, 2, 0, 0, 1, 1, 0, 0, 2, 0, 0, 0,
0, 0, 2, 2, 2, 0, 2, 1, 2, 1, 0, 2, 0, 2, 2, 2, 2, 2, 0, 0, 2, 1,
1, 0, 1, 0, 2, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 2, 0, 2, 1, 1, 2, 0,
2, 2, 1, 0, 2, 2, 2, 2, 0, 2, 0, 0, 1, 1, 1, 2, 1, 0, 2, 0, 2, 0,
0, 0, 1, 0, 2, 2, 2, 0, 1, 0, 1, 2, 1, 0, 0, 0, 2, 2, 0, 1, 0, 2,
0, 2, 0, 0, 0, 2, 0, 2, 2, 0, 1, 1, 0, 0, 0, 0, 2, 2, 0, 1, 2, 0,
2, 0, 2, 2, 0, 2, 1, 2, 0, 2, 0, 0, 0, 0, 2, 0, 2, 1, 0, 0, 0, 0,
2, 1, 1, 0, 1, 2, 0, 2, 0, 1, 2, 2, 0, 0, 0, 0, 2, 2, 2, 1, 0, 2,
2, 0, 0, 0, 1, 0, 0, 0, 0, 0, 2, 2, 0, 0, 0, 2, 0, 0, 0, 0, 0, 2,
0, 0, 2, 0, 0, 2, 1, 0, 0, 0, 2, 0, 0, 2, 0, 1, 2, 2, 0, 2, 2, 0,
2, 0, 0, 0, 2, 2, 0, 0, 2, 0, 0, 0, 0, 0, 2, 1, 2, 2, 1, 2, 1, 1,
2, 0, 0, 2, 1, 2, 2, 0, 0, 0, 0, 2, 0, 1, 0, 1, 0, 2, 2, 2, 0, 1,
0, 0, 2, 0, 0, 0, 0, 2, 2, 0, 0])
df['cluster']=y_predicted
df
| Name | Age | Fare | Initial | cluster | |
|---|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 0.271174 | 0.014151 | Mr | 2 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 0.472229 | 0.139136 | Mrs | 0 |
| 2 | Heikkinen, Miss. Laina | 0.321438 | 0.015469 | Miss | 0 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 0.434531 | 0.103644 | Mrs | 0 |
| 4 | Allen, Mr. William Henry | 0.434531 | 0.015713 | Mr | 0 |
| ... | ... | ... | ... | ... | ... |
| 886 | Montvila, Rev. Juozas | 0.334004 | 0.025374 | Other | 0 |
| 887 | Graham, Miss. Margaret Edith | 0.233476 | 0.058556 | Miss | 2 |
| 888 | Johnston, Miss. Catherine Helen "Carrie" | 0.271174 | 0.045771 | Miss | 2 |
| 889 | Behr, Mr. Karl Howell | 0.321438 | 0.058556 | Mr | 0 |
| 890 | Dooley, Mr. Patrick | 0.396833 | 0.015127 | Mr | 0 |
891 rows × 5 columns
km.cluster_centers_
array([[0.40370991, 0.04801379],
[0.64396415, 0.11667009],
[0.20482494, 0.05977556]])
df1=df[df.cluster==0]
df2=df[df.cluster==1]
df3=df[df.cluster==2]
plt.scatter(df1.Age,df1['Fare'],color = 'green')
plt.scatter(df2.Age,df2['Fare'],color = 'red')
plt.scatter(df3.Age,df3['Fare'],color = 'black')
plt.xlabel('Age')
plt.ylabel('Fare')
plt.legend()
No handles with labels found to put in legend.
<matplotlib.legend.Legend at 0x1be66036e88>

df1=df[df.cluster==0]
df2=df[df.cluster==1]
df3=df[df.cluster==2]
plt.scatter(df1['Fare'],df1.Age,color = 'green')
plt.scatter(df2['Fare'],df2.Age,color = 'red')
plt.scatter(df3['Fare'],df3.Age,color = 'black')
plt.ylabel('Age')
plt.xlabel('Fare')
plt.legend()
No handles with labels found to put in legend.
<matplotlib.legend.Legend at 0x1be65296a08>

k_rng=range(1,10)
sse=[]
for k in k_rng:
km=KMeans(n_clusters=k)
km.fit(df[['Age','Fare']])
sse.append(km.inertia_)
sse
[33.163249933113455,
18.859245724888236,
13.110890535996486,
8.754991374324431,
6.378184765822988,
5.303403333391413,
4.352438977975344,
3.759949542306482,
3.227046378111474]
plt.xlabel('K')
plt.ylabel('Sum of squared error')
plt.plot(k_rng,sse)
[<matplotlib.lines.Line2D at 0x1be651dcd08>]

Hierarchical Clustering
import numpy as np
from matplotlib import pyplot as plt
import pandas as pd
importing data:
df=pd.read_csv('Unsupervised.csv')
df
| Name | Age | Fare | |
|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 22.0 | 7.2500 |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 | 71.2833 |
| 2 | Heikkinen, Miss. Laina | 26.0 | 7.9250 |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 | 53.1000 |
| 4 | Allen, Mr. William Henry | 35.0 | 8.0500 |
| ... | ... | ... | ... |
| 886 | Montvila, Rev. Juozas | 27.0 | 13.0000 |
| 887 | Graham, Miss. Margaret Edith | 19.0 | 30.0000 |
| 888 | Johnston, Miss. Catherine Helen "Carrie" | NaN | 23.4500 |
| 889 | Behr, Mr. Karl Howell | 26.0 | 30.0000 |
| 890 | Dooley, Mr. Patrick | 32.0 | 7.7500 |
891 rows × 3 columns
df.isnull().sum()
Name 0
Age 177
Fare 0
dtype: int64
df['Initial']=0
for i in df:
df['Initial']=df.Name.str.extract('([A-Za-z]+)\.') #extracting Name initials
pd.crosstab(df.Initial,train_data.Sex).T.style.background_gradient(cmap='summer_r')
df['Initial'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess',
'Jonkheer','Col','Rev','Capt','Sir','Don'],['Miss',
'Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr'],inplace=True)
df.groupby('Initial')['Age'].mean()
df.loc[(df.Age.isnull()) & (df.Initial=='Mr'),'Age']=33
df.loc[(df.Age.isnull()) & (df.Initial=='Mrs'),'Age']=36
df.loc[(df.Age.isnull()) & (df.Initial=='Master'),'Age']=5
df.loc[(df.Age.isnull()) & (df.Initial=='Miss'),'Age']=22
df.loc[(df.Age.isnull()) & (df.Initial=='Other'),'Age']=46
df.Age.isnull().any()
False
df.head()
| Name | Age | Fare | Initial | |
|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 22.0 | 7.2500 | Mr |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 | 71.2833 | Mrs |
| 2 | Heikkinen, Miss. Laina | 26.0 | 7.9250 | Miss |
| 3 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 | 53.1000 | Mrs |
| 4 | Allen, Mr. William Henry | 35.0 | 8.0500 | Mr |
X= df.iloc[:,[1,2]].values
X
array([[22. , 7.25 ],
[38. , 71.2833],
[26. , 7.925 ],
...,
[22. , 23.45 ],
[26. , 30. ],
[32. , 7.75 ]])
Using dendrogram to find the optimal number of clusters
import scipy.cluster.hierarchy as sch
dendrogram = sch.dendrogram(sch.linkage(X, method = 'ward'))

Fitting hierarchical clustering to the dataset
from sklearn.cluster import AgglomerativeClustering
AgglomerativeClustering()
hc=AgglomerativeClustering(affinity='euclidean', linkage='ward')
y_hc= hc.fit_predict(X)
plt.scatter(X[y_hc==0,0], X[y_hc==0,1], s = 100, c= 'red', label='cluster 1')
plt.scatter(X[y_hc==1,0], X[y_hc==1,1], s = 100, c= 'blue', label='cluster 2')
plt.scatter(X[y_hc==2,0], X[y_hc==2,1], s = 100, c= 'green', label='cluster 3')
plt.title('Clusters of Passengers')
plt.xlabel('Age')
plt.ylabel('Fare')
plt.legend()
plt.show()

The End
from IPython.display import Image
Image(url="https://d13ezvd6yrslxm.cloudfront.net/wp/wp-content/images/titanic-jack-rose-door.jpg")

